Methyl Ferulate Induced Conformational Changes of DeOxyHbS: Implication on Sickle Erythrocyte Polymerization
Abstract
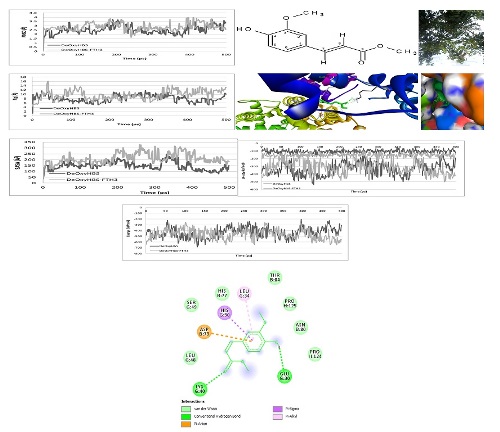
Sickle cell disease (SCD) is a molecular disease caused by substituting glutamic acid with valine at the β-6 position of the hemoglobin, leading to the polymerization of erythrocytes that contain the hemoglobin afterward leads to severe clinical consequences. Polymerization of sickle hemoglobin occurs only in the deoxygenated form i.e only sickle deoxyhemoglobin (DeOxyHbS) polymerizes. SCD is predominant in children living in Africa, especially in West Africa. Therefore, molecular docking and molecular dynamic simulation studies were carried out on methyl ferulate isolated from Ficus thonningii leaves, a known antisickling plant used in Eastern Nigeria to manage SCD. The Harborne procedure was used for extraction, whereas the combination of column chromatography and flash chromatography was used for the isolation and purification of active principles of the leaves extract. The structure of methyl ferulate was determined based on nuclear magnetic resonance (NMR) analysis. A binding affinity of -5.8 kcal/mol indicated that methyl ferulate binds to DeOxyHbS and could interfere with the processes that trigger sickle hemoglobin polymerization in vitro. The observed variations in perturbation of both DeOxyHbS and FTH3-DeOxyHbS complex root mean square deviation (RMSD), the radius of gyration (Rgyr), solvent accessible surface area (SASA), potential energy (PE), and Van der Waal’s (VDW) interactions were obtained from the molecular dynamic simulation studies of the binding site amino acid residue performed at 500 ps and suggest that in silico methyl ferulate binds with amino acid residues reported being involved in sickle hemoglobin polymerization and thus may possess antisickling potentials in vitro.
Full Text:
PDFReferences
N.L. Yambeau, P.C. Biapa Nya, C.A Pieme, K.D. Tchouane, C.B. Kengne, Fotsing, P.J. Nya Nkwikeu, A.F. Feudjio, P.B. Telefo, Ethnopharmacological study of the medicinal plants used in the treatment of sickle cell anemia in the west region of Cameroon, Evid. Based Complement. Altern. Med., 2022, 5098428.
. W. A. Eaton, J. Hofrichter, S Hemoglobin Gelation and Sickle Cell Disease, Blood., 1987, 70, 1245–1266.
W. A. Eaton, J. Hofrichter, Sickle Cell Hemoglobin Polymerization, Adv Protein Chem., 1990, 40, 63–79.
D.C. Rees, T.N. Williams, M.T. Gladwin, Sickle-Cell Disease, Lancet., 2010, 376, 2018–2031.
R.E. Ware, M. de Montalembert, L. Tshilolo, M.R. Abboud, Sickle Cell Disease, Lancet., 2017, 390, 311–323.
D.J. Harrington, K. Adachi, W.E. Royer, The High-Resolution Crystal Structure of Deoxyhemoglobin S, J Mol Biol., 1997, 272, 398–407.
E.A. Padlan, W.E. Love, Refined Crystal Structure of Deoxyhemoglobin S II. Molecular Interactions in the Crystal, J Biol Chem., 1985, 260, 8280.
E.A. Padlan, W.E. Love, Refined Crystal Structure of Deoxyhemoglobin S I. Restrained Least-Squares Refinement at 3.0-8 Resolution, J Biol Chem., 1985, 260, 8272.
O.E. Nnodu, H.A. Isa, R.I. Chianumba, Y. Tanko, J.H. Iyobosa, J.H. Nnebe-Agumadu, A. Sopekan, C. Ohiaeri, A. Adeniran, G. Shedul, O. Owolabi, A.D. Adekile, O.I. Olopade, F.B. Piel, Implementing newborn screening for sickle cell disease as part of immunization programs in Nigeria: a feasibility study, Lancet Haematol., 2020, 7, e534-e540.
B.P. Schoenborn, Dichloromethane as an antisickling agent in sickle cell hemoglobin (gas binding to hemoglobin/sickle cell disease), Proc Natl Acad Sci., 1976, 73, 4195-4199.
J.T. Finch, M.F. Perutz, J.F. Bertles, J. Dobler, Structure of sickle erythrocyte and of sickle-cell hemoglobin fibers, Proc Natl Acad of Science, USA., 1973, 70, 718-722.
R. Josephs, H.S. Jarosch, S.J. Edelstein, Polymorphism of sickle cell hemoglobin fibers, J Mol Biol., 1976, 102, 409-426.
B. Magdoff-Fairchild, P.H. Swerdlow, J.F. Bertles, Intermolecular organization of deoxygenated sickle hemoglobin determined by x-ray diffraction, Nature, 1972, 239, 217-219.
B.P. Schoenborn, Binding of Xenon to Horse Haemoglobin, Nature, 1965, 208, 760-762.
M.F. Perutz, G. Fermi, D.J. Abraham, C. Poyart, E. Bursaux, Hemoglobin as a Receptor of Drugs and Peptides: X-Ray Studies of the Stereochemistry of Binding, J Amer Chem Soc., 1986, 108, 1064–1078.
K. Adachi, T. Asakura, Demonstration of a Delay Time during Aggregation of Diluted Solutions of Deoxyhemoglobin S and Hemoglobin CHarlem in Concentrated Phosphate Buffer, J Biol Chem., 1978, 253, 6641–6643.
K. Adachi, T. Asakura, Nucleation-Controlled Aggregation of Deoxyhemoglobin S. Possible Difference in the Size of Nuclei in Different Phosphate Concentrations, J Biol Chem., 1979, 254, 7765–7771.
K. Adachi, T. Asakura, Kinetics of the Polymerization of Hemoglobin in High and Low Phosphate Buffers. Blood Cells., 1982, 8, 213–224.
F.A. Ferrone, J. Hofrichter, W.A. Eaton, Kinetics of Sickle Hemoglobin Polymerization. I. Studies Using Temperature-Jump and Laser Photolysis Techniques, J Mol Biol., 1985, 183, 591–610.
F.A. Ferrone, J. Hofrichter, W.A. Eaton, Kinetics of Sickle Hemoglobin Polymerization. II. A Double Nucleation Mechanism, J Mol Biol., 1985, 183, 611–631.
O. Galkin, W. Pan, L. Filobelo, R.E. Hirsch, R. Nagel, P.G. Vekilov, Two-Step Mechanism of Homogeneous Nucleation of Sickle Cell Hemoglobin Polymers, Biophys J., 2017, 93, 902–913.
A. Horwich, Protein Aggregation in Disease: A Role for Folding Intermediates Forming Specific Multimeric Interactions, J Clin Invest., 2002, 110, 1221–1232.
C.A. Ross, M.A. Poirier, Protein aggregation and neurodegenerative disease, Nat Med., 2004, 10, S10–S17.
P. Ball, Water Is an Active Matrix of Life for Cell and Molecular Biology, Proc Natl Acad Sci, USA., 2017, 201703781.
M.C. Bellissent-Funel, A. Hassanali, M. Havenith, R. Henchman, P. Pohl, F. Sterpone, D. van der Spoel, Y. Xu, A.E. Garcia, Water Determines the Structure and Dynamics of Proteins, Chem Rev., 2016, 116, 7673–7697.
V.M.A. Ingram, Specific Chemical Difference Between the Globins of Normal Human and Sickle-Cell Anamia Hamoglobin, Nature, 1956, 178, 792–794.
M. Murayama, Molecular Mechanism of Red Cell “Sickling.”, Science, 1966, 153, 145–149.
C.T. Noguchi, A.N. Schechter, Sickle Hemoglobin Polymerization in Solution and Cells, Annu Rev Biophys Biophys Chem., 1985, 14, 239–263.
L.L. Pauling, H.A. Itano, S.J. Singer, I.C. Wells, Sickle Cell Anemia, a Molecular Disease. Science, 1949, 110, 543.
J.A. McCammon, S.C. Harvey, Dynamics of Proteins and Nucleic Acids, Cambridge University Press, Cambridge, 1987, 260, 8272-8279.
H. Muirhead, J.M. Cox, L. Mazzarella, M.F. Perutz, Structure and function of hemoglobin: III. A Three dimensional Fourier synthesis of human deoxyhemoglobin at 5.5 Å resolution, J Mol Biol., 1976, 28, 117-156.
N. Galamba, S. Pipolo, On the Binding Free Energy and Molecular Origin of Sickle Cell Hemoglobin Aggregation, J Phys Chem., 2018, 2-30.
K.I. Ijoma, V.I.E. Ajiwe, C.O. Alisa, Antimicrobial Analysis and Structural Elucidation of Active Compounds of Nauclea latifolia Stem Extract (Pin Cushion Tree), Amer J Chem Appl., 2017, 4, 21-26.
K.I. Ijoma, V.I.E. Ajiwe, C.I. Awuzie, Antimicrobial Analysis and Structural Elucidation of Active Compounds of Dialium indum leaves extract (Velvet Tarmarind), Int J Pharm Chem., 2016, 6, 237-244.
K.I. Ijoma, V.I.E. Ajiwe, Phytochemical Screening of Dialium indum leaf extract (Velvet Tarmarind), Int J Phytopharm., 2017, 7, 6-13.
Y. Roskov, G. Ower, T. Orrell, D. Nicolson, N. Bailly, P.M. Kirk, T. Bourgoin, R.E. Dewalt, W. Decock, E. Nieukerken, L. Penev, Specie 2000, Naturalis, Leiden, the Netherland, 2020.
Global Biodiversity Information Facility Secretariat. Ficus thonningii Blume. GBIF Backbone Taxonomy, Checklist dataset, 2019.
J.B. Harbone, Phytochemical Methods- A guide to modern Techniques of plant analysis, 3rd edition, London, Chapman and Hall, 1998, 6-7.
F.C. Bernstein, T.F. Koetzle, G.J. Williams, E.F. Meyer, M.A. Jr., M.D. Brice, J.R. Rodgers, O. Kennard, T. Shimanouchi, M. Tasumi, The protein data bank: a computer-based archival file for macromolecular structures. J Mol Biol., 1977, 25, 535-542.
O. Trott, A.J. Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading, J Comput Chem., 2010, 31, 455-461.
M.D. Hanwell, D.E. Curtis, D.C. Lonie, T. Vandermeersch, E. Zurek, G.R. Hutchison, Avogadro: An advanced semantic chemical editor, visualization, and analysis platform, J Cheminform., 2012, 4, 17
L. Ferreira, R. dos Santos, G. Oliva, A.D. Andricopulo, Molecular docking and structure-based drug design strategies, Molecules, 2015, 20, 13384–13421.
D.S. Godsell, G.M. Morris, A.J. Olson, Automated docking of flexible ligands: Applications of AutoDock, J Mol Recogn., 1996, 9, 1–5.
G.M. Morris, R. Huey, A.J. Olson, Using autodock for ligand-receptor docking, Curr Protoc Bioinform., 2008, 24, 8–14.
Biovia Dassault Systemes. Discovery studio visualizer 2020. San Diego, CA, USA.
. W.L. Delano, Pymol: An open-source molecular graphics tool. CCP 4 Newsletter on Protein crystallography, 2002, 40, 82-92.
J.C. Philips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, C.R.D. Skeel, L. Kale, K. Schulten, Scalable molecular dynamics with NAMD, J Comput Chem., 2005, 26, 1781-1802.
R.B. Best, X. Zhu, J. Shim, E.M. Lopes, J. Mittal, M. Feig, A.D. Mackerell, Jr, Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone φ, ψ and Side-Chain χ1 and χ2 Dihedral Angles, J Chem Theory Comput., 2012, 8, 3257–3273.
M.A. Jr, M. Feig, B.C. Rd, Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations, J Comput Chem., 2004, 25, 1400–1415.
M.F. Harrach, B. Drossel, Structure and dynamics of TIP3P, TIP4P, and TIP5P water near smooth and atomistic walls of different hydroaffinity, J Chem Phys., 2014, 140, 3393–3393.
T. Schlick, Molecular Modeling and Simulation, Springer, 2002, 1-162.
D.S. Cerutti, R.E. Duke, T.A. Darden, T. P. Lybrand, Staggered Mesh Ewald: an extension of the smooth particle-mesh Ewald method adding great versatility, J Chem Theory Comput., 2009, 5, 2322.
T. Darden, D. York, L. Pedersen, Particle mesh Ewald: An Nṡlog (N) method for Ewald sums in large systems. J Chem Phys, 1993, 98,
–10092.
U. Essmann, L. Perera, M.L. Berkowitz, T. Darden, H. Lee, G.L. Pedersen, A smooth particle mesh Ewald method, J Chem Phys., 1995, 103, 8577–93.
G. Bussi, D. Donadio, M. Parrinello, Canonical sampling through velocity rescaling, J Chem Phys., 2007, 126, 014101.
M. Parrinello, A. Rahman, Polymorphic transitions in single crystals: a new molecular dynamics method, J Appl Phys., 1981, 52,
–90.
B. Hess, H. Bekker, H.J.C. Berendsen, J.G.E.M. Fraaije, LINCS: A Linear Constraint Solver for molecular simulations, J Comput Chem., 1997, 18, 1463–72.
W. Humphrey, A. Dalke, K. Schulten, VMD: visual molecular dynamics, J Mol Graph., 1996, 14, 33–38.
A. Tanaka, A. Kato, T. Tsuchiya, Isolation of Methyl ferulate From Rice Bran Oil, J Amer Oil Chem Soc., 1971, 48, 95-97.
A. Ilmiawati, D. Anggraini, G. Syahbirin, D.U. Rahayu, D. U. P. Sugita, Methyl Ferulate from Methanol Extract of Indonesian Sausage Fruit (Kigelia Africana). The 8th International Conference of the Indonesian Chemical Society (ICICS), AIP Conference Proceedings, 2000, 2243, 030009-1–030009-5.
N.T.M., Phuong, T.T. Cuong, D.N. Quang, Anti-inflammatory activity of Methyl Ferulate isolated from Stemona tuberosa Lour, Asian Pac J Trop Med., 2014, 7(Suppl 1), S327-S331.
M.F. Perutz, Mechanisms regulating the reactions of human hemoglobin with oxygen and carbon monoxide Annu Rev Physiol., 1990, 52, 1-25.
R.H. Garrett, C.M. Grisham, Biochemistry, 4th edition, Cengage learning, 2013.
M.K. Safo, O. Abdulmalik, R. Danso-Danquah, J.C. Burnett, S. Nokuri, G.S. Joshi, F.N., Musayev, T. Asakura, D.J. Abraham, Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds, J Med Chem., 2004, 47, 4665–4676.
M.R. Waterman, K. Yamaoka, L. Dahm, J. Taylor, G.L. Cottam, Noncovalent modification of deoxyhemoglobin S and erythrocyte sickling. Proc Natl Acad Sci USA, 1974, 71, 2222-2226.
M.F. Perutz, Stereochemistry of Cooperative Effects in Haemoglobin: Haem–Haem Interaction and the Problem of Allostery, Nature., 1970, 228, 726–734.
P.D. Ross, J. Hofrichter, W.A. Eaton, Calorimetric and optical characterization of sickle cell hemoglobin. J Mol Biol., 1975, 96, 239-256.
P.D. Ross, J. Hofrichter, W.A. Eaton, Thermodynamics of gelation of sickle cell hemoglobin, J Mol Biol., 1977, 115, 111-134.
M.K. Safo, M.H. Ahmed, M.S. Ghatge, T. Boyiri, Hemoglobin-Ligand binding: Understanding Hb Function and Allostery on atomic level, Biochim Biophys Acta., 2011, 1814, 797–809.
M.Y. Lobanov, N. Bogatyreva, O. Galzitskaya, Radius of gyration as an indicator of protein structure compactness, Mol Biol (Mosk)., 2008, 42, 623–628.
NOTES
Biovia Discovery studio homepage can be accessed at https://discover.3ds.com/discovery-studio-visualizer-download
Information on NAMD program can be found at www.ks.uiuc.edu/Research/namd/
Avogadro homepage www.avogadro.cc
Information on Autodock tools and vina script can be accessed using www.autodock.scripps.edu
VMD home page www.ks.uiuc.edu/Research/vmd
Chemsketch homepage www.acdlabs.com/resources/freeware/chemsketch/index.php
More information on CHARMM GUI can be found at www.charmm-gui.org
Information on the PDB file format can be found at www.rcsb.org/structure/2HBS
Microsoft windows environment and Excel can be found at www.microsoft.com
DOI: http://dx.doi.org/10.13171/mjc02208061631ijoma
Refbacks
- There are currently no refbacks.
Copyright (c) 2022 Mediterranean Journal of Chemistry
