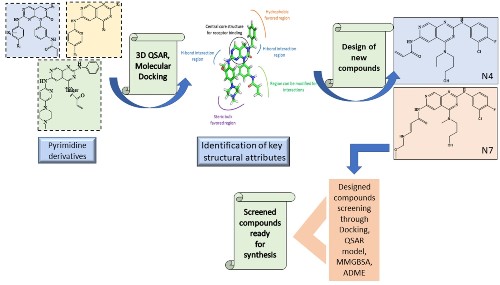
Structural identification of novel pyrimidine derivatives as epidermal growth factor receptor inhibitors using 3D QSAR, molecular docking, and MMGBSA analysis: a rational approach in anticancer drug design
Abstract
Full Text:
PDFReferences
R.L. Siegel, K.D. Miller, A. Jemal, Cancer statistics, 2016. CA: a cancer journal for clinicians, 2016, 66(1), 7-30.
S.V. Sharma, D.W. Bell, J. Settleman, D.A. Haber, Epidermal growth factor receptor mutations in lung cancer, Nature Reviews Cancer, 2007, 7(3), 169-181.
M.A. Olayioye, The ErbB signaling network: receptor heterodimerization in development and cancer, The EMBO journal, 2000, 19(13), 3159-3167.
M.M. Moasser, Targeting the function of the HER2 oncogene in human cancer therapeutics. Oncogene, 2007, 26(46), 6577-6592.
C.E. Geyer, Lapatinib plus capecitabine for HER2-positive advanced breast cancer, New England Journal of Medicine, 2006, 355(26), 2733-2743.
S.Chang, Design, synthesis, and biological evaluation of novel conformationally constrained inhibitors targeting epidermal growth factor receptor threonine790→ methionine790 mutant, Journal of medicinal chemistry, 2012, 55(6), 2711-2723.
T.S. Mok, Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. New England Journal of Medicine, 2009, 361(10), 947-957.
F.A. Shepherd, Erlotinib in previously treated non–small-cell lung cancer, New England Journal of Medicine, 2005, 353(2), 123-132.
K.D. Carey, Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Research, 2006, 66(16), 8163-8171.
M.L. Sos, Chemogenomic profiling provides insights into the limited activity of irreversible EGFR Inhibitors in tumor cells expressing the T790M EGFR resistance mutation, Cancer Research, 2010, 70(3), 868-874.
Kim, Y., et al., The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor, Molecular cancer therapeutics, 2012, 11(3), 784-791.
Z. Song, Challenges and perspectives on the development of small-molecule EGFR inhibitors against T790M-mediated resistance in non-small-cell lung cancer: miniperspective, Journal of medicinal chemistry, 2016, 59(14), 6580-6594.
H. Patel, Recent updates on third-generation EGFR inhibitors and emergence of fourth-generation EGFR inhibitors to combat C797S resistance. European Journal of Medicinal Chemistry, 2017, 142, 32-47.
X. Ji, Design, synthesis and biological evaluation of novel 6-alkenylamides substituted of 4-anilinothieno [2, 3-d] pyrimidines as irreversible epidermal growth factor receptor inhibitors, Bioorganic & medicinal chemistry, 2014, 22(7), 2366-2378.
H. Zhang, Discovery of 2, 4, 6-trisubstitued pyrido [3, 4-d] pyrimidine derivatives as new EGFR-TKIs, European Journal of Medicinal Chemistry, 2018, 148, 221-237.
B. Bhardwaj, Insight into structural features of phenyltetrazole derivatives as ABCG2 inhibitors for the treatment of multidrug resistance in cancer, SAR and QSAR in Environmental Research, 2019, 30(7), 457-475.
Y. Jian, Molecular modeling study for the design of novel peroxisome proliferator-activated receptor gamma agonists using 3D-QSAR and molecular docking, International Journal of Molecular Sciences, 2018, 19(2), 630.
A. Dixit, Development of CoMFA, advance CoMFA and CoMSIA models in pyrroloquinazolines as thrombin receptor antagonist, Bioorganic & medicinal chemistry, 2004, 12(13), 3591-3598.
S. Bhattacharya, Comparative Molecular Field Analysis on 4 (3H) Quinazolinone derivatives for the Development of Potential Antimicrobial Agents, Journal of Drug Delivery and Therapeutics, 2019, 9(2), 603-611.
A. Golbraikh, A. Tropsha, Beware of q2! Journal of molecular graphics and modelling, 2002, 20(4), 269-276.
K.S. Gajiwala, Insights into the aberrant activity of mutant EGFR kinase domain and drug recognition, Structure, 2013, 21(2), 209-219.
S. Agarwal, A. Dixit, S.K. Kashaw, Ligand and structure-based virtual screening of chemical databases to explore potent small molecule inhibitors against breast invasive carcinoma using recent computational technologies, Journal of Molecular Graphics and Modelling, 2020, 107591.
M. Mishra, Integrated computational investigation to develop molecular design of quinazoline scaffold as promising inhibitors of plasmodium lactate dehydrogenase, Journal of Molecular Structure, 2020, 1207, 127808.
A. Daina, O. Michielin, V. Zoete, SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules, Scientific reports, 2017, 7, 42717.
J.S. Delaney, Predicting aqueous solubility from structure, Drug Discovery Today, 2005, 10(4), 289-295.
I. Moriguchi, Simple method of calculating octanol/water partition coefficient, Chemical and pharmaceutical bulletin, 1992, 40(1), 127-130.
C.A. Lipinski, Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced drug delivery reviews, 1997, 23(1-3), 3-25.
A.K. Ghose, V.N. Viswanathan, J.J. Wendoloski, A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases, Journal of combinatorial chemistry, 1999, 1(1), 55-68.
Y.C. Martin, A bioavailability score, Journal of medicinal chemistry, 2005, 48(9), 3164-3170.
DOI: http://dx.doi.org/10.13171/mjc02304141691kashaw
Refbacks
- There are currently no refbacks.
Copyright (c) 2023 Mediterranean Journal of Chemistry
